The research team of Prof. Morigen and Associate Prof. Fan Lifei who are with IMU publishes its research article in mLife online in collaboration with Prof. Kirsten Skarsta who is with the University of Oslo. The research finds that DnaA might regulate the transcription of the structural gene hisG, but not that of the leader gene hisL; DnaA might interact with the HisL‐SL RNA through rDnaA boxes to strengthen the transcription attenuation of his operon; and the mechanism seems to be phylogenetically conserved in Gram‐negative bacteria. The article proposes a DnaA-dependent riboswitch for transcription attenuation of the his operon.

The previous research on the model of the transcription attenuation indicates that, in the presence of a specific amino acid, transcription will terminate following the formation of the attenuator stem‐loop, whereas in the absence of the same amino acid, the translating ribosome will stall on the leader mRNA9, thereby inhibiting attenuator formation and allowing the synthesis of the respective amino acid. However, it had not been proved that the DnaA protein was involved in the regulation of transcription attenuation. The current literature shows that the model can be applied to the regulation of transcription attenuation of operons of tryptophane, phenylalanine, threonine, leucine and histidine.
1. DnaA regulates the transcription attenuation of his operon through its interaction with HisL-SL RNA.
The research studies the molecular mechanism for transcription attenuation with the his operon of Gram bacteria. It is found that the transcription attenuation of his operon is dependent on DnaA and HIS l-SL(hisL-stem loop region) and RNA; and DnaA might regulate the transcription of the structural gene hisG, but not that of the leader gene hisL. Then on the basis of this finding, with RNAIP(immunoprecipitation) it is found that HisL-SLRNA and DnaA are coprecipitated while HisL-SLRNA and gapA-RNA are not(Fig. 1A). Interestingly, rDnaA-boxes1,2,3,4 are identified in HisL-SL RNA while rDnaA-box is not identified in gapA-RNA-the same result with RNA IP and the HisL‐SL RNA and rDnaA boxes are phylogenetically conserved in Gram‐negative bacteria. The lack of RDnaA-boxes1,2 and 3 weakens the interaction of HisL-SL RNA with DnaA to improve the transcription of his operon while the lack of rDnaA-box4 does not influence the interaction with DnaA and the transcription of his operon(Fig.1C, D).
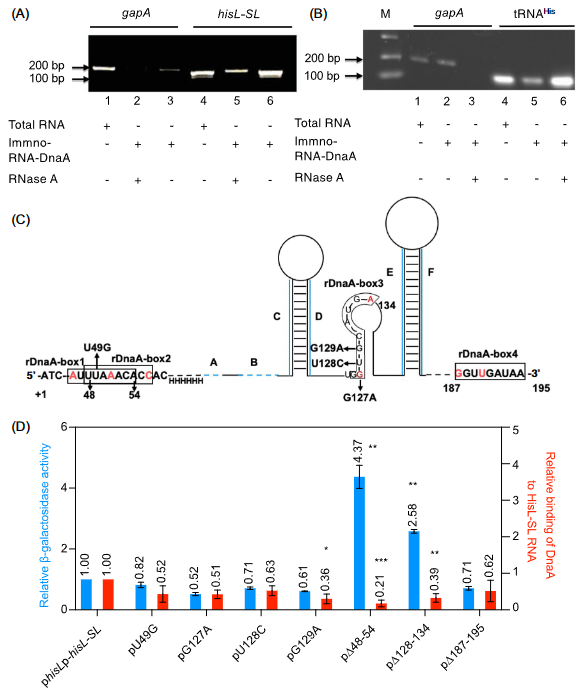
Fig. 1 DnaA interacts with HisL-SL RNA and lack of rDnaA-boxes1,2,3 weakens the interaction and transcription attenuation.
2. Excess tRNAHis strengthens the transcription attenuation of the his operon, which is dependent on DnaA and HisL-SL RNA.
It is found in the research that excess tRNAHis reduces the transcription of the his operon in wt MOR1378 cells and the ΔhisL, ΔhisL‐SL, dnaAA345S mutant derivatives(Fig. 2A), which indicates that excess tRNAHis strengthens the attenuation of the his operon . In the ΔhisL‐SL dnaAA345S and ΔhisL dnaAA345S double mutants, excess tRNAHis exerts no prominent effect on his transcription (Fig.2A), which demonstrates that repression of tRNAHis is dependent on DnaA and His-SL RNA. The destroyed Shine Dalgarno sequence in 5' region of hisL gene, the destroyed start codon of the gene and the inserted stop codon in front of the His codon, will all strengthen the attenuation of his operon(Fig. 2B,C). To sum up, the research proposes a model of a DnaA-dependent riboswitch for transcription attenuation of the his operon(Fig.2D). The model assumes that the binding of DnaA to rDnaA boxes in regulatory RNA of the his operon ensures the formation of the attenuator EF and vice versa. And ribosome loading and tRNAHis loading play a role in transcription attenuation of the his operon.
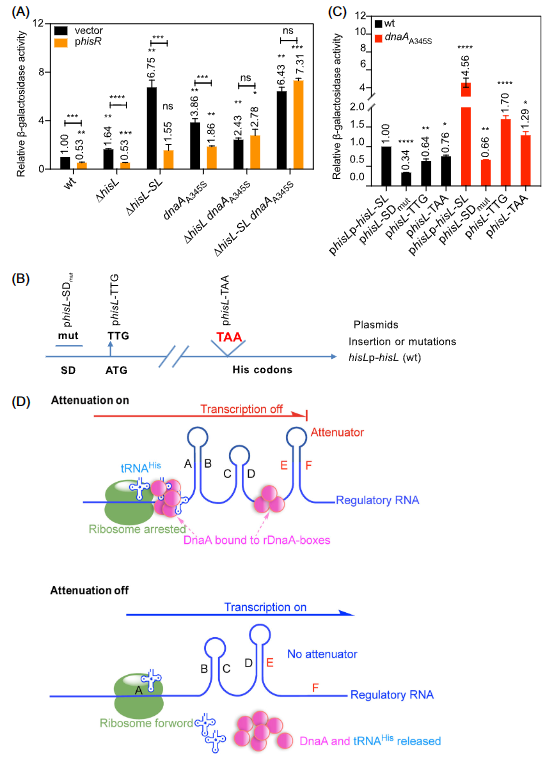
Fig.2 Excess tRNAHis strengthens the transcription attenuation of the his operon.
A model for a DnaA-dependent and HisL-SL RNA-DnaA-mediated riboswitch for transcription attenuation
Currently, the transcription attenuation mechanism in the textbook Molecular Biology assumes that the interaction of mRNA and regulatory RNA with ribosome changes the stem-loop structure of RNA and forms or eliminates transcription attenuator; and a specific amino acid concentration plays a role of riboswitch. However, the research shows that the interaction of DnaA protein with regulatory RNA of the his operon plays a role of “switch”. More interestingly, in the medium without exogenous amino acid, the addition of histidine does not change the transcription of the his operon and the reason for this needs to be further explored.
How to cite this article: Yao Y, Sun H, Wurihan, Gegeheng, Gezi, Skarstad K, et al. A DnaA‐dependent riboswitch for transcription attenuation of the his operon. mLife. 2023; 2: 126–140.
URL: https://doi.org/10.1002/mlf2.12075

First author: Yao Yuan, associate research fellow
Institution of the first author: Inner Mongolia University(IMU)
Introduction to the first author: Yao is supervisor of master students of the School of Life Sciences of IMU and supervisor of master students of Inner Mongolia Medical University, executive council member of Inner Mongolia Society of Biochemistry and Molecular Biology and vice chairman of Youth Committee of Neurology Department Branch of Inner Mongolia Society of Doctors and focuses on the studies of mechanism of microorganism to adapt to the surroundings and pathogenesis of cerebral apoplexy.

Corresponding author: Prof. Morigen
Institution of the corresponding author: School of Life Sciences of IMU
Introduction to corresponding author: Morigen, doctor and supervisor of PhD students, are the members of the editorial board of Chinese Journal of Cell Biology and Chinese Journal of Biochemistry and Molecular Biology. He focuses on the studies of the duplication of DNA of colon bacillus, genome stability and regulation of genetic expression and has led the research of 6 projects supported by the National Natural Science Foundation of China and published over 60 academic articles. He has discovered and revealed the novel DnaA-dependent riboswitch of transcription attenuation, the mechanism of DnaA to regulate the duplication, damage and genetic expression of DNA and the new model of spatial regulation for genetic expression.

Corresponding author: Associate Prof. Fan Lifei
Institution of the corresponding author: School of Life Sciences of IMU
Introduction to corresponding author: Fan Lifei, doctor and supervisor of master students, is vice chairman of Inner Mongolia Society of Biochemistry and Molecular Biology. She focuses on the studies of the duplication of DNA and tumorigenesis and the small G protein of Rho and migration and infiltration of tumor. She has led the research of 2 projects supported by the National Natural Science Foundation of China, published over 30 academic articles in such journals as J Autoimmunity and mLife and revealed for the first time the molecular mechanism through which the small G protein-Rif gets involved in congenital immune response of cells.