The team of Prof. Zuo Yongchun of IMU have recently achieved the latest progress in protein sequence , protein structure and protein function analysis in the field of bioinformatics with their thesis entitled RaacFold: a webserver for 3D visualization and analysis of protein structure by using reduced amino acid alphabets published in Nucleic Acids Research (influence factor: 16.971), an internationally renowned academic journal. Published by Oxford University Press, a globally renowned publisher, Nucleic Acids Research concerns itself with physical, chemical, biochemical and biological studies of nucleic acid and protein involved in metabolism and interactions of nucleic acid and enjoys authoritative influence in the field of studies of nucleic/protein and bioinformatics in the world.
Protein is the direct reflector of the genetic features of cells and the entire organisms. Acclaimed by academicians of the two Chinese Academies as one of the top ten progresses in science and technology in the world in 2020 and listed by Science as the world’s No. 1 breakthrough in science in 2021, AlphaFold, an AI instrument for the prediction of protein structure offers a solution to the great challenges in the field of biology over the past 50 years. The AI instrument greatly accelerates the process of health monitoring, precise medicine and R & D of new medicines for mankind and offers the latest frontier technology in the studies of the structure prediction and function analysis of proteins for grassland livestock and other large-sized animals. The greater complexity of protein analysis compared with those of other biomacromolecules will generally impair computing efficiency and increase information redundancy and possibilities of excess matching for computing biologists. This in turn will usually impair the interpretation, interaction and biological annotation of protein functions for experimental biologists. To minimize the influence of the defects mentioned above, theoretically it is possible to compress the alphabets consisting of 20 natural amino acids with similar biochemical features and frequency distributions into simplified amino acid alphabets consisting of fewer amino acid categories so as to simplify the sequence and structure analysis of proteins, which is the theoretical basis for us to develop RaacFold, a protein analysis platform.
The thesis published in the journal
By making use of 687 amino acid alphabets extracted from 58 reduction and calculations methods, three analysis instruments are developed by RaacFold: Protein Analysis, Align Analysis and Multi Analysis. Protein Analysis and Align Analysis, which provide sequences and structures with similar and simplified characteristics, can identify the function regions distributed in protein structures more clearly to obtain key information of characteristics. Subsequently the designing of artificial protein is simplified and redundant information is avoided. In addition, by allowing users to explore the protein structure and physical mutation and conservativeness in the function evolution, the Multi Analysis provides essential information for identifying and exploring non-paralogs functions of paralogs. In the interactive interface of RaacFold, users’ real-time edition, rendering and exporting of sequences, structures and function annotation are supported. The platform provides four view modes: natural coding mode, reduced coding mode, evolutionary conservation mode and reliability prediction mode. To create a direct visual effect of protein structure, the RaacFold provides powerful 2D and 3D rendering functions which support shades, silhouette, roughness and other details for adjusting rendering parameters. Meanwhile the display of different layers of protein structure, including surface and bulk (molecule surface, Gaussian cube, etc), secondary structure (cartoon, ribbon, etc) and atoms (ball and sticks, space-filling, etc) is supported.
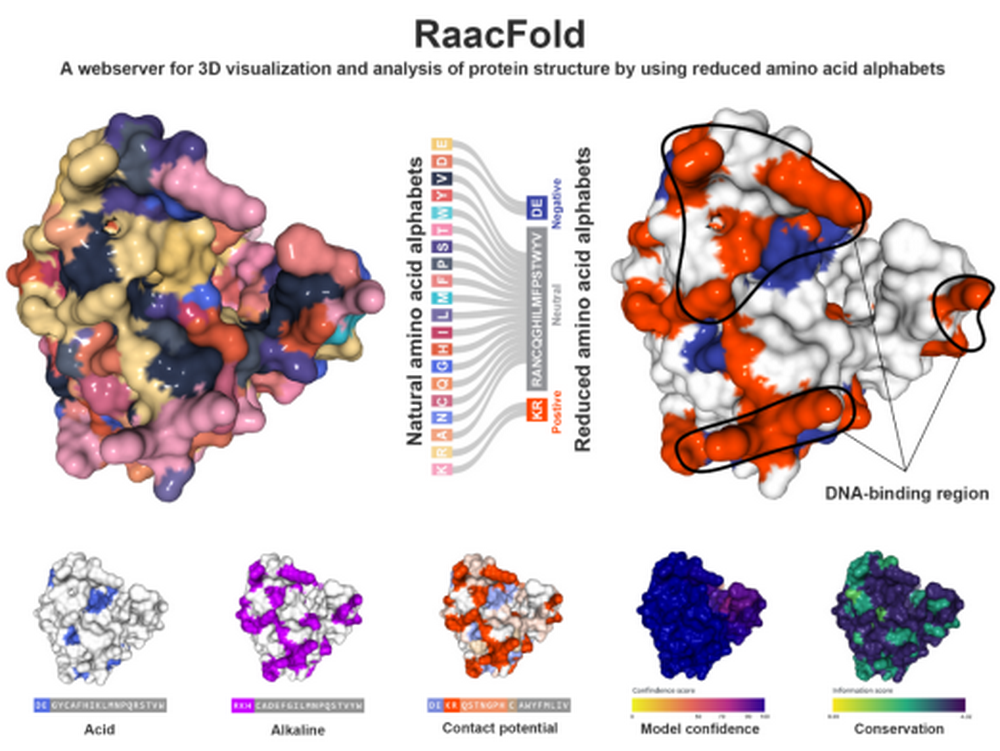
Diagram abstract of RaacFold
RaacFold offers a sound solution to the high characteristic dimension and redundancy in the processing of protein sequence, structure and function analysis by AI and performs extraordinarily in reducing protein complexity and capturing the conservative characteristics hidden in noise signals. For computing and experimental biologists, it may become an important part of protein analysis which includes homology testing, evolution inference and function prediction. The platform can provide far-sighted theories in studies of the functioning mechanism of protein, offer strong guidance for studies in development and evolution and furnish a highly efficient platform of analysis for protein design. Before publishing the thesis, the team of Prof. Zuo Yongchun had built PseKRAAC, a platform for reduced analysis and feature extraction of protein sequence amino acid (Bioinformatics. 2017; ESI 1% highly cited thesis globally), RaacBook, a database of reduced protein amino acid (Database, 2019; software copyright registration No. 2019SR0467812) and RaacLogo, a platform for visual analysis of the function domain of protein sequence (Briefings in Bioinformatics. 2021; ESI 1% highly cited thesis globally).

The team of Prof. Zuo Yongchun
A brief introduction to the corresponding author: Professor Zuo Yongchun, PhD supervisor, graduated from the School of Physical Science and Technology of IMU with Doctor’s degree in Biological Physics in 2011, specializing in bioinformatics. Prof. Zuo currently holds the positions of member of the Board of Experts of the Chinese Society for Cell Biology, corresponding member of the Board of Experts of China Computer Federation, vice Secretary General of Inner Mongolia Biophysics and Bioinformatics Society, editorial board member of Current Gene Therapy and topics guest editor of Frontiers. Prof. Zuo published more than 70 theses as the corresponding or first author in world-renowned journals including Nucleic Acids Research, Cellular and Molecular Life Sciences, Briefings in Bioinformatics and Bioinformatics, which have been cited for more than 1900 times (Google, h index: 27) by academicians of the National Academy of Sciences of the United States and scholars from other countries. Six theses of Prof. Zuo are listed as ESI 1% highly cited theses globally. So far Prof. Zuo has obtained about a dozen national invention patents and software copyrights. Prof. Zuo is currently in charge of 5 projects sponsored by the National Natural Science Foundation of China, projects sponsored by the Foundation for Cultivating Excellent Youth in Natural Science of Inner Mongolia , Programs for Young Scientific Talents of Universities and Colleges, Programs for Key Technologies, and is listed in the Grassland Talents of Inner Mongolia and its subsequent supporting programs, 321 Talents Project of the New Century and its subsequent supporting programs, etc.
The project is jointly undertaken by the School of Life Sciences of IMU, the State Key Laboratory of Reproductive Regulation & Breeding of Grassland Livestock and Inner Mongolia University of Science & technology. The thesis has IMU as the organization of the first author and the first corresponding author, Zheng Lei, PhD Candidate admitted in 2019 as the first author, Prof. Zuo Yongchun as the first corresponding author and Prof. Xing Yongqiang from the School of Life Science & Technology of Inner Mongolia University of Science & Technology as the corresponding co-author. The research is sponsored by the National Natural Science Foundation of China (62061034, 62171241, 61861036, 62161039), the Foundation for Cultivating Excellent Youth in Natural Science of Inner Mongolia (2017JQ04), Programs for Key Technologies of Inner Mongolia (2021GG0398) and Programs for Young Talents of Science & Technology (NJYT-18-B01).
URL for the thesis: https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkac399/6593113

